Analyzing Data
The Analyze class contains functions which can be used to identify cohorts in datasets, perform differential expression and pathway analysis, and execute meta-analysis workflows.
Parameters:
-
token(str, default:None) –Authentication token from polly
Usage
from polly.analyze import Analyze
analysis = Analyze(token)
identify_cohorts
This function is used to get the cohorts that can be created from samples in a GEO dataset. Please note: Currently only Bulk RNASeq datasets from GEO source are supported. If results are generated for other datatypes or datasource, they may be inaccurate. If you want to use this functionality for any other data type and source, please reach out to polly.support@elucidata.io
Parameters:
-
repo_key(int / str) –repo_id or repo_name in str or int format
-
dataset_id(str) –dataset_id of the GEO dataset. eg. "GSE132270_GPL11154_raw"
Returns:
-
DataFrame–Dataframe showing values of samples across factors/cohorts.
run_meta_analysis
Use this function to execute the Polly DIY Meta-Analysis Pipeline. Only the 'geo_transcriptomics_omixatlas' omixatlas is supported currently.
Parameters:
-
repo_key(int / str) –repo_id or repo_name in str or int format
-
workspace_id(int) –the workspace in which the datasets and results should be stored
-
analysis_name(str) –name of your analysis, eg. "MA_BRCA_run1". The reports, datasets, and results will be stored in a folder with this name.
-
design_formulas(dict) –key:value pair of atleast 2 dataset ids and a list of design formulas. eg. dataset_id:[design formula control, design formula perturbation]
-
samples_to_remove(list, default:[]) –List of samples to omit, if any. Defaults to [].
Examples
# Install polly python
!sudo pip3 install polly-python --quiet
# Import libraries
from polly import analyze
AUTH_TOKEN=(os.environ['POLLY_REFRESH_TOKEN'])
analysis = analyze.Analyze(AUTH_TOKEN)
Identify Cohorts
Factors based on which cohorts can be created for this dataset: ['genotype', 'gender', 'age__at_the_time_of_sacrifice_', 'treatment', 'experiment', 'curated_genetic_modification_type']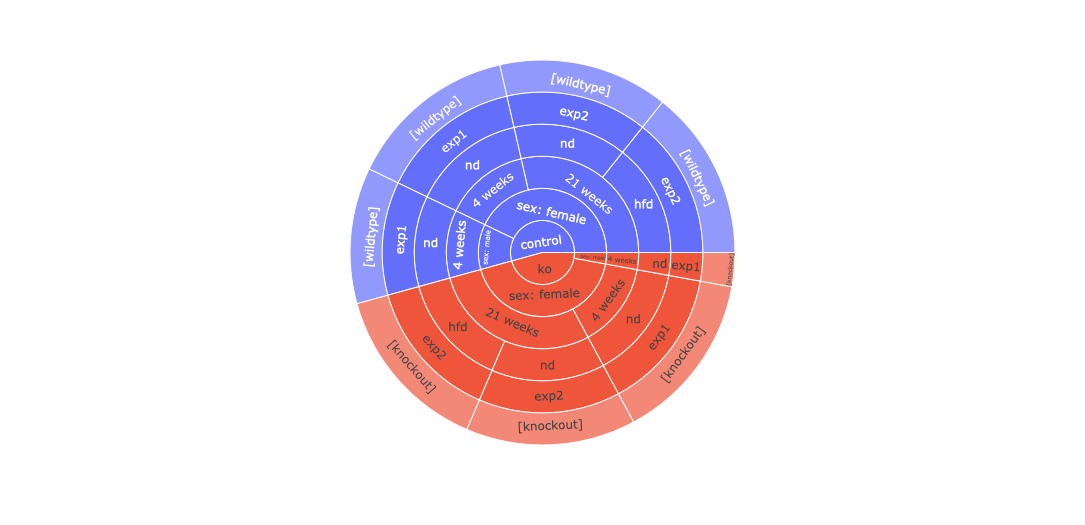
| genotype | gender | age__at_the_time_of_sacrifice_ | treatment | experiment | curated_genetic_modification_type | |
|---|---|---|---|---|---|---|
| geo_accession | ||||||
| GSM3301731 | ko | sex: female | 4 weeks | nd | exp1 | [knockout] |
| GSM3301732 | ko | sex: female | 4 weeks | nd | exp1 | [knockout] |
| GSM3301736 | control | sex: male | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301737 | control | sex: male | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301739 | control | sex: female | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301742 | ko | sex: female | 21 weeks | nd | exp2 | [knockout] |
| GSM3301744 | ko | sex: female | 21 weeks | nd | exp2 | [knockout] |
| GSM3301749 | control | sex: female | 21 weeks | hfd | exp2 | [wildtype] |
| GSM3301750 | control | sex: female | 21 weeks | nd | exp2 | [wildtype] |
| GSM3301753 | control | sex: female | 21 weeks | hfd | exp2 | [wildtype] |
| GSM3301754 | control | sex: female | 21 weeks | nd | exp2 | [wildtype] |
| GSM3301757 | ko | sex: female | 21 weeks | hfd | exp2 | [knockout] |
| GSM3301758 | ko | sex: female | 21 weeks | hfd | exp2 | [knockout] |
| GSM3301759 | ko | sex: female | 21 weeks | hfd | exp2 | [knockout] |
| GSM3301760 | ko | sex: female | 21 weeks | hfd | exp2 | [knockout] |
| GSM3301761 | ko | sex: female | 21 weeks | nd | exp2 | [knockout] |
| GSM3301727 | control | sex: male | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301728 | ko | sex: male | 4 weeks | nd | exp1 | [knockout] |
| GSM3301729 | ko | sex: female | 4 weeks | nd | exp1 | [knockout] |
| GSM3301730 | control | sex: female | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301733 | ko | sex: female | 4 weeks | nd | exp1 | [knockout] |
| GSM3301734 | ko | sex: female | 4 weeks | nd | exp1 | [knockout] |
| GSM3301735 | control | sex: female | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301738 | control | sex: male | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301740 | control | sex: female | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301741 | control | sex: female | 4 weeks | nd | exp1 | [wildtype] |
| GSM3301743 | ko | sex: female | 21 weeks | nd | exp2 | [knockout] |
| GSM3301745 | ko | sex: female | 21 weeks | nd | exp2 | [knockout] |
| GSM3301746 | control | sex: female | 21 weeks | hfd | exp2 | [wildtype] |
| GSM3301747 | control | sex: female | 21 weeks | hfd | exp2 | [wildtype] |
| GSM3301748 | control | sex: female | 21 weeks | hfd | exp2 | [wildtype] |
| GSM3301751 | control | sex: female | 21 weeks | nd | exp2 | [wildtype] |
| GSM3301752 | control | sex: female | 21 weeks | nd | exp2 | [wildtype] |
| GSM3301755 | control | sex: female | 21 weeks | nd | exp2 | [wildtype] |
| GSM3301756 | ko | sex: female | 21 weeks | hfd | exp2 | [knockout] |
# If you want to plot a sunburst on specific columns of interest, please use the following code:
import plotly.express as px
metadata = identify_cohorts(repo_key, dataset_id)
fig = px.sunburst(metadata, path=['column_1','column_2','column_n'])
fig.show()
Run Meta-Analysis
repo = "bulkrnaseq_staging_oa"
datasets = ['GSE144269_GPL24676_raw','GSE114564_GPL11154_raw','GSE77314_GPL9052_raw']
designformula = {'GSE144269_GPL24676_raw' :
[{'tumor_non_tumor':'non-tumor'}, {'tumor_non_tumor':'tumor'}],
'GSE114564_GPL11154_raw' :
[{'curated_disease':'[Normal]'}, {'curated_disease':'[Carcinoma, Hepatocellular]'}],
'GSE77314_GPL9052_raw' :
[{'curated_control':'0'}, {'curated_control':'1'}]
}
ws_id= 14164
analysis_name = 'HCC_test1'
| Job ID | Job Name | Job State | |
|---|---|---|---|
| 0 | cb4074a13e464f3282310be2f5c60845 | Polly DIY pipeline: Meta Analysis | SUCCEEDED |
| Job ID | Job Name | Job State | |
|---|---|---|---|
| 0 | 553b1df3fcff41c4a91b15ecbb582633 | Polly DIY pipeline: Meta Analysis | SUCCEEDED |
A new version of Polly CLI is available. To update, execute the command npm update -g @elucidatainc/pollycli
Success: Logged in to Polly as shreya.raghavendra@elucidata.io
================================== POLLY LOGIN SUCCESS =======================================
“=============================== GIT CLONING POLLY-PYTHON ===================================”
Cloning into 'polly-python-code'...
“================================== GIT CLONING SUCCESS =======================================”
A new version of Polly CLI is available. To update, execute the command npm update -g @elucidatainc/pollycli
polly://HCC_test1_cohorts.csv
./cohort.csv
Success: Download complete
==================================== EXECUTING JOBS ==========================================
downloading data file:GSE77314_GPL9052_raw.gct: 100%|██████████| 29.4M/29.4M [00:00<00:00, 69.9MiB/s]
downloading data file:GSE144269_GPL24676_raw.gct: 100%|██████████| 34.3M/34.3M [00:01<00:00, 33.7MiB/s]
downloading data file:GSE114564_GPL11154_raw.gct: 100%|██████████| 58.7M/58.7M [00:01<00:00, 32.8MiB/s]
[1] "reading read_design_formula_csv "
[1] "done reading read_design_formula_csv "
[1] "process_gct_files started ....."
[1] "processing GCT files..."
parsing as GCT v1.3
datasets/GSE114564_GPL11154_raw.gct 62548 rows, 106 cols, 8 row descriptors, 40 col descriptors
parsing as GCT v1.3
datasets/GSE144269_GPL24676_raw.gct 29565 rows, 140 cols, 1 row descriptors, 42 col descriptors
parsing as GCT v1.3
datasets/GSE77314_GPL9052_raw.gct 35831 rows, 100 cols, 2 row descriptors, 42 col descriptors
================================= EXECUTING JOBS SUCCESS =====================================
==================================== SYNC RESULTS TO POLLY - STARTING ==========================================
A new version of Polly CLI is available. To update, execute the command npm update -g @elucidatainc/pollycli
Success: Sync complete
A new version of Polly CLI is available. To update, execute the command npm update -g @elucidatainc/pollycli
Success: Sync complete
================================= SYNC RESULTS TO POLLY - SUCCESS =====================================
Tutorial Notebooks
Identifying Cohorts for Multi-Factor Experiments From data findability, identifying cohorts, to running Meta-analysis